Where did chromosome 19 got its gene clusters?. SMBE Conference. Yokohama, Japan
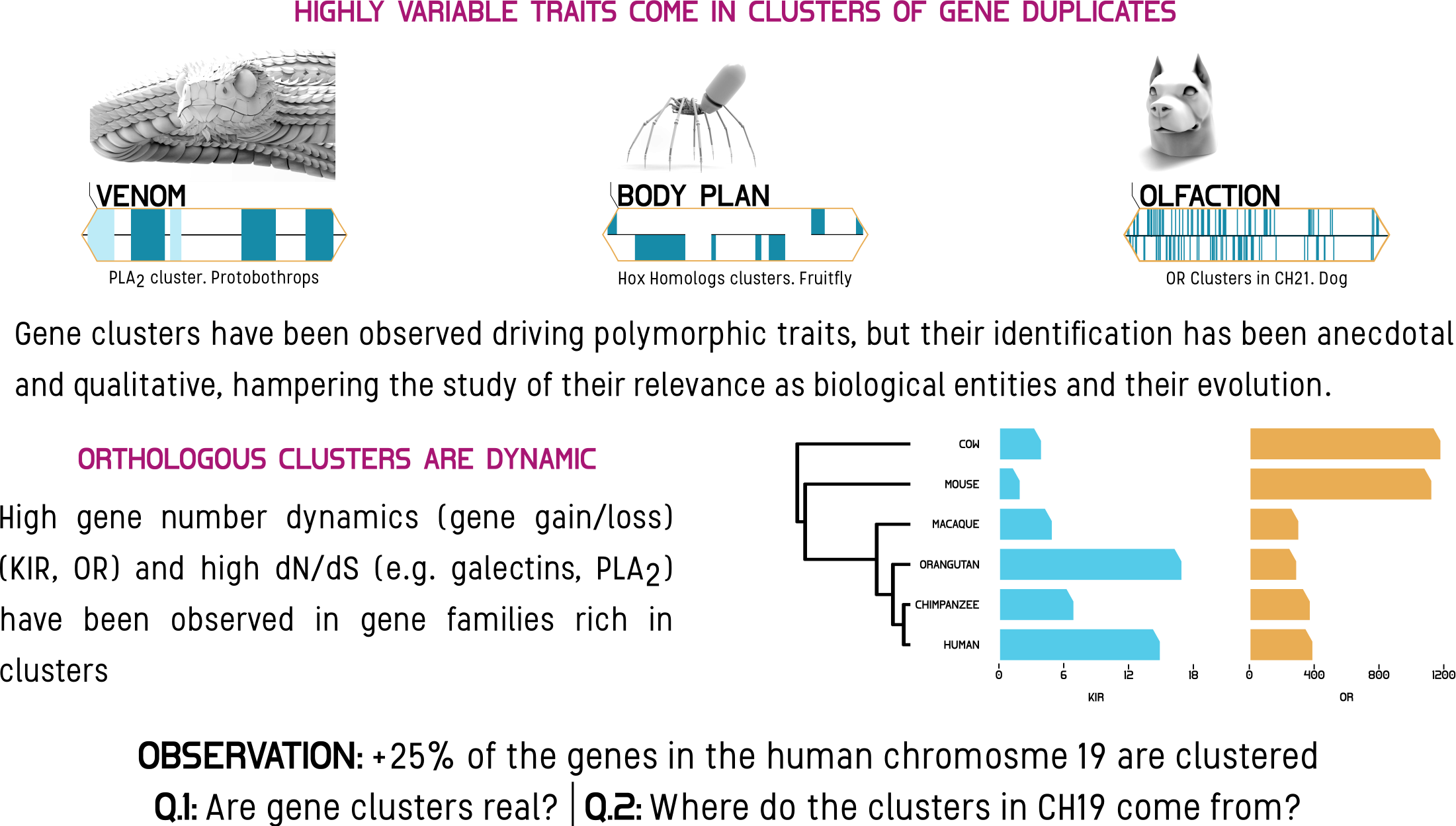
Clusters of duplicated genes (CTDGs) are great contributors to the diversity exhibited by many phenotypes, like snake venom, animal body plans, and olfaction. To systematically study CTDGs, we developed CTDGFinder, a homology-based algorithm that statistically takes into account the size and spacing of duplicate gene locations in a genome, and has been shown to identify several well-known mammalian CTDGs (e.g., the Hox, globin, and protocadherin CTDGs). We found that more than 20% of human genes belong to CTDGs and that CTDGs span 7% of the total length of the human genome. Moreover, the distribution of CTDGs is not uniform across chromosomes, with chromosome 19 being the most significantly enriched in CTDGs. To investigate how the remarkable density of CTDGs in human chromosome 19 evolved, we compared human chromosome 19 to its orthologous chromosomes in other mammalian genomes. We found that chromosome 19 evolved from a chromosome fusion event prior to the split of the primate and rodent lineages. Interestingly, only one of the two chromosome 19 ancestors shows a history of conserved high CTDG density across mammals. Furthermore, genomic alignments of chromosome 19 orthologous regions show that its ancestral chromosomes experienced contraction, increasing chromosome 19 CTDG density. These results highlight the importance of CTDGs in chromosomal evolution, and open avenues of research towards a deeper understanding of the interactions between CTDGs and molecular evolutionary processes.