Quests and adventures in Genomic Geography. Three Minutes Thesis Competition. Winner. Vanderbilt University. Department of Biological Sciences
Link to full Oral presentation
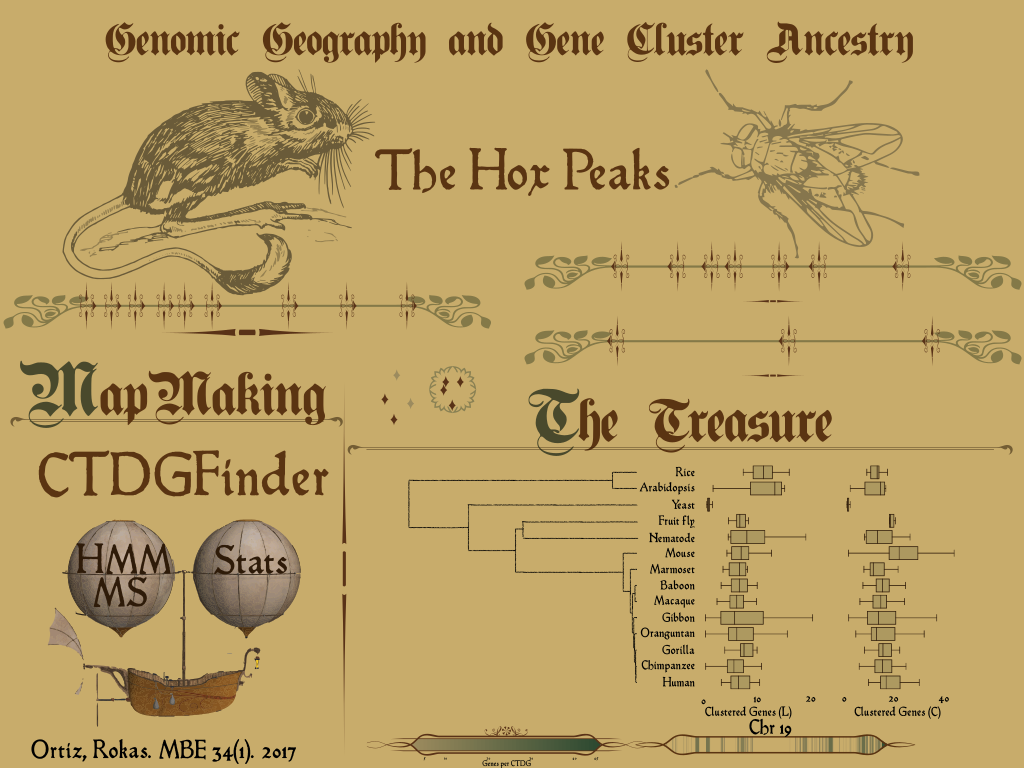
The genome, that wonderful land. We, humble explorers, have cataloged many genes, many hills. Even more, as we mapped genes in different genomes, we discovered that many genes come in clusters of duplicated genes! Those mountain ranges, spanning large sections of our genomes, provide a lush environment for diverse phenotypes: olfaction, immunity, and body plan architecture are just some examples. Here, the maps of a set of hox genes important in body plan in mouse and fly are shown. Although both sets of genes come from the same ancestor, they are very different. We can see how one compact cluster in mouse, is broken apart in two in the fly, we can see that the distance between genes is so different. In other words, how different are those clusters? What can those clusters tell us about the history of the animal genomes?
You see, many of the wonders of the genome have been studied in isolation, saving and drawing maps one genome at a time. That approach proved very useful in a time when those maps were traced by hand, and walked by foot. If we want to study the genomes of many species, we need to lift ourselves from the ground, and fly over those genomes, recording their characteristics from the same altitude, with the same measuring stick.
In order to accomplish that, I designed a ship called CTDGFinder. A computational algorithm to identify gene clusters, to be exact. Using sequence homology, computer vision, statistics, now we are able to examine how genomes from different species differ in their gene clusters composition. This is how I plan to do genomic geography with this ship. Because I am using the same procedure to identify gene clusters, I can make comparisons between clusters, and because I am using homology information, I will be able to group clusters of the same origin.
With that information, I have discovered that many genomes of different sizes have a similar proportion of clustered genes, but I have also noticed, that some chromosomes have different distribution of clustered genes, like the case of the human chromosome 19, in which more than 30% of its genes are clustered.
In the future, I will continue my journey on this ship. I will visit different clusters on mammals and plants, in which extensive cartography has been made. With the wealth of information available, I will test if significant variations on cluster compositions correspond to changes in evolutionary parameters, giving us clues regarding the genomic processes that can accelerate or deccelerate evolution
