Juan Felipe Ortiz
10 years experience designing HPC and cloud-native scalable solutions to run NGS and comparative genomics analyses, from standardized to complex, in production environments.
Awards
Social Media Presence
Publications
This has been an arduous path, but it is satisfying to see the products of my work, big and small.
Oral presentation - A Novel Homology-Based Algorithm for Identifying Closely Spaced Clusters of Tandemly Duplicated Genes. Quest for Orthologs 5. Los Angeles, USA.
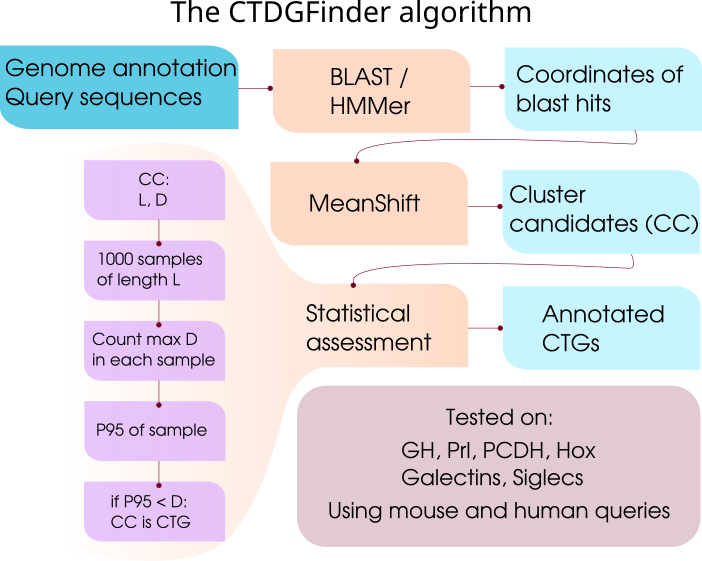
Poster - Where did chromosome 19 got its gene clusters?. SMBE Conference. Yokohama, Japan
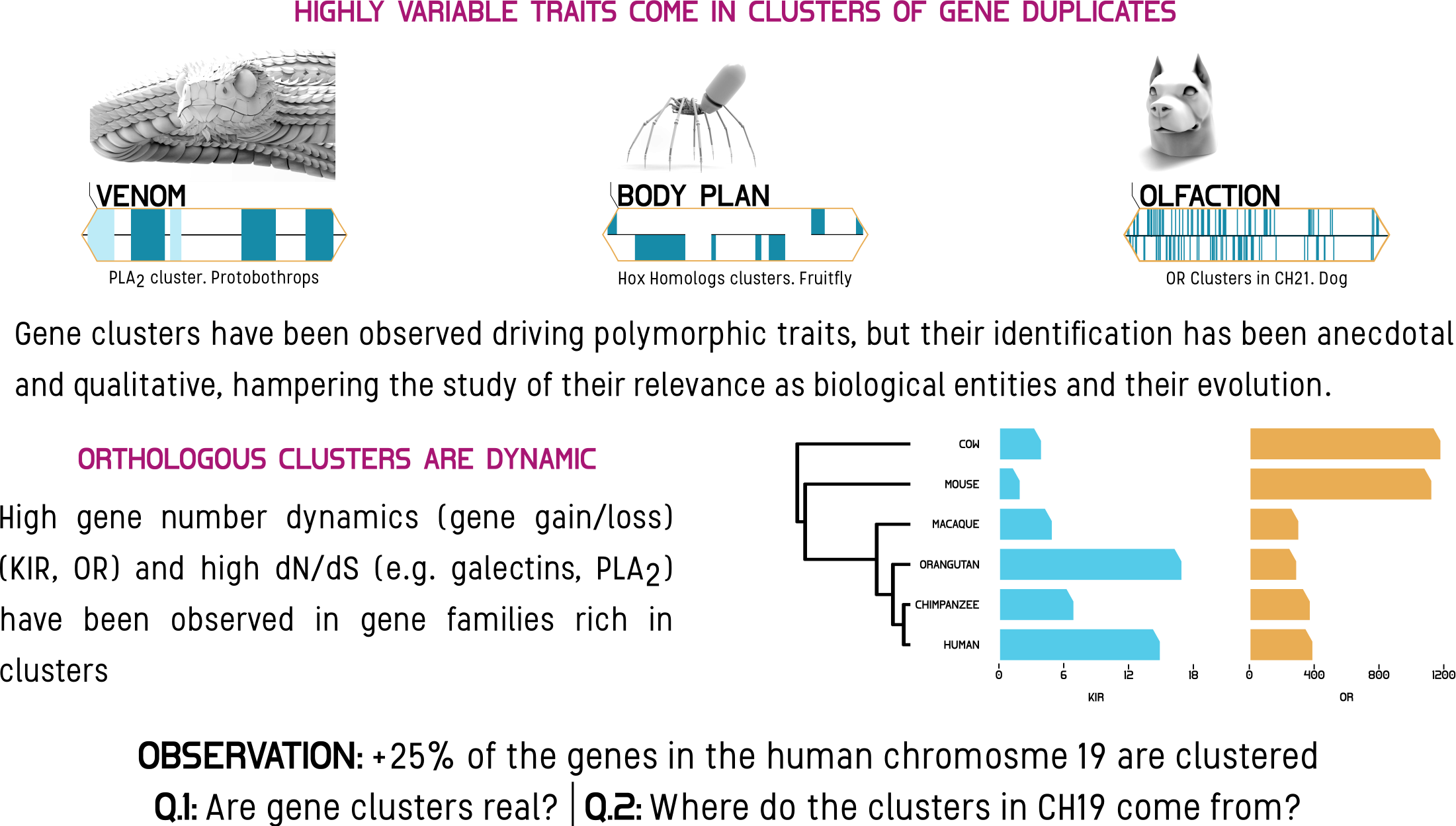
Oral presentation - Quests and adventures in Genomic Geography. Three Minutes Thesis Competition. Winner. Vanderbilt University. Department of Biological Sciences
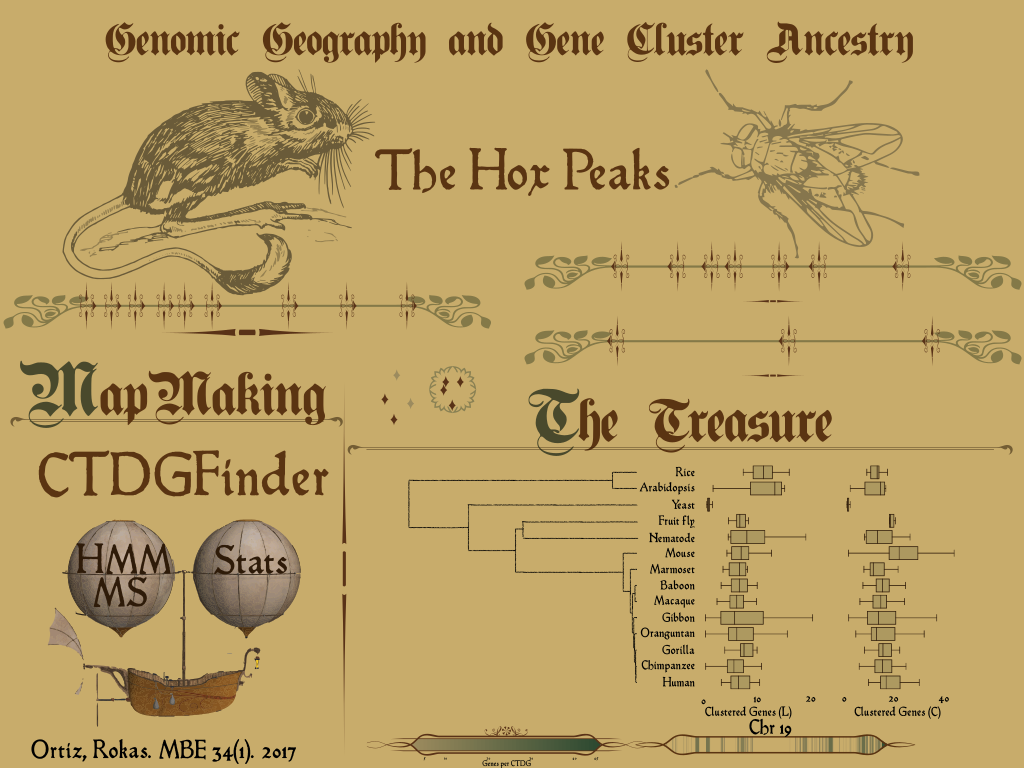
Article - CTDGFinder: A Novel Homology-Based Algorithm for Identifying Closely Spaced Clusters of Tandemly Duplicated Genes
Article - A genetic map of cassava ( Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes
Article - Identification of Immunity-related Genes in Arabidopsis and Cassava Using Genomic Data
Article - Genomic Survey of Pathogenicity Determinants and VNTR Markers in the Cassava Bacterial Pathogen Xanthomonas axonopodis pv. Manihotis Strain CIO151
Article - Did the prion protein become vulnerable to misfolding after an evolutionary divide and conquer event?
Article - Rapid Evolutionary Dynamics of Structural Disorder as a Potential Driving Force for Biological Divergence in Flaviviruses
Article - Notes on the marine algae of the International Biosphere Reserve Seaflower, Caribbean Colombia II: diversity of drift algae in San Andres island, Caribbean Colombia.
Experience
Lead Bioinformatics Engineer
- Design cloud-based (AWS) architecture for deploying and running scalable NGS workflows.
- Encode scalable and optimized NGS workflows using the WDL and Nextflow languages.
- Design full-stack web application for submitting and tracking NGS workflows.
- Design and execute research projects and collaborative initiatives in NGS science.
Special postoctoral fellow
- Develop algorithms for the assessment of novel chromatin conformation capture experiments.
- Design and execute research projects on comparative chromatin dynamics and their association with ageing.
Postdoctoral fellow
- Develop data analysis pipelines and downstream analysis for RNA-seq experiments.
- Design and execute research programs combining structural genomics and evolutionary biology methods.
- Deploy a genome browser integrating multi-omics data for the brownbanded bamboo shark genome.
Education
Vanderbilt University
Publication: Ortiz, J F Rokas, A . (2016). CTDGFinder: A Novel Homology-Based Algorithm for Identifying Closely Spaced Clusters of Tandemly Duplicated Genes. Molecular Biology and Evolution 34 (1) : 215-229.
University of Wyoming
MacDonald, M L Masterson, P Uversky, V N Siltberg-Liberles, J . (2013). Rapid Evolutionary Dynamics of Structural Disorder as a Potential Driving Force for Biological Divergence in Flaviviruses. Genome Biology and Evolution 5 (3) : 504-513.
National University of Colombia
Publication: Ortiz, J F Gavio, B . (2010). Notes on the marine algae of the International Biosphere Reserve Seaflower, Caribbean Colombia II: diversity of drift algae in San Andres island, Caribbean Colombia. Caribbean Journal of Science 46 (2-3) : 313-321.